
The GloWPa model
The GloWPa (Global Waterborne Pathogen) model simulates emissions and concentrations of pathogens (currently Cryptosporidium and rotavirus) to/in surface water. These pathogens are known to be a leading cause of diarrhoeal diseases globally. GloWPa focuses on human and livestock emissions of pathogens that end up in surface water systems through various pathways. Special attention is paid to the storage and removal of pathogens in manure storage facilities or wastewater treatment systems. The concentration of the pathogens is calculated using the human and livestock emissions and information hydrology. The Global Waterborne Pathogen (GloWPa) model has been developed over the years and we constantly work to improve the model. The model The following publications provide the best available overview of the model:
- Hofstra et al. 2013 is the first version of the model and explores opportunities to model Cryptosporidium at a global scale.
- Vermeulen et al. 2015 look further at the human Cryptosporidium emissions, adding the population that does not have access to sewers (except for people with pit latrines and septic tanks), specifically for Bangladesh and India.
- Kiulia, Hofstra et al. 2015 apply the Vermeulen et al (2015) model improvements to the world to estimate Rotavirus emissions.
- Hofstra and Vermeulen 2016 use the Kiulia et al (2015) model for human Cryptosporidium emissions to surface water in a scenario analysis, studying changes in population, urbanisation, sanitation and waste water treatment. The paper concludes that waste water treatment is essential when sewers are put in place.
- Vermeulen et al 2017 provide a strongly improved model for the livestock emissions to land.
- Vermeulen et al 2019 provide simulated Cryptosporidium concentrations in rivers worldwide. The paper also shows a first validation of the model.
- Okaali et al 2021 developed the GloWPa model for rotavirus concentrations in Uganda. They have added on-site emissions to the model.
- Okaali et al 2022 use a new version of the human emission model that includes all 13 sanitation categories that the Joint Monitoring Project of WHO and UNICEF also includes. This version of the model has also been used in the Pathogen Flow and Mapping Tool.
- In the WaterPath project the GloWPa model code is currently professionalised and will be developed into an R package.
Schematic of the most recent version of the human emission model.
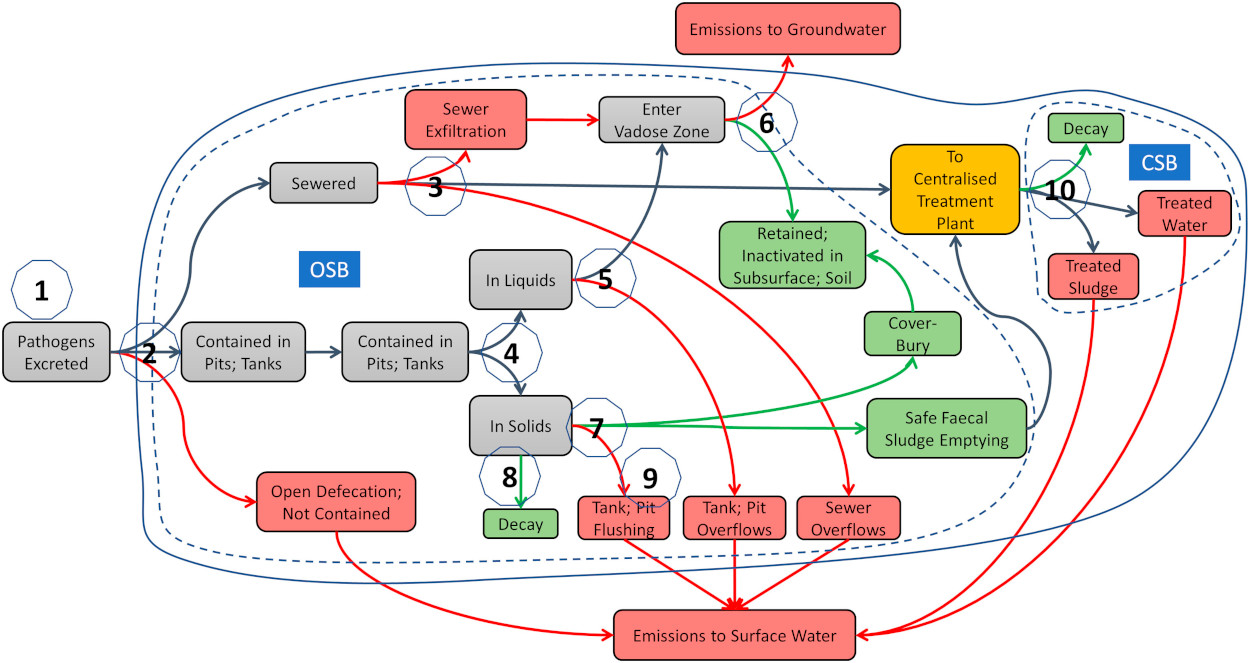
Schematic of the livestock emission model.

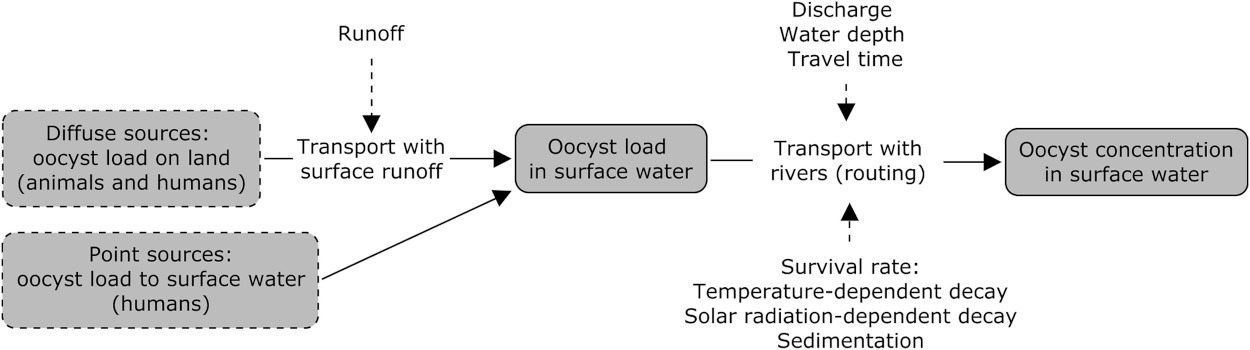
Schematic of the concentration model.
Data request
We are always looking for measured microorganisms data for the validation of the GloWPa model, the first spatially explicit global model of diarrheal pathogens in surface waters (see below for an overview of the model). Relevant datasets are records of Cryptosporidium, rotavirus, E. coli or other microorganism concentrations (or counts and volume, if possible with recovery rates) in rivers and streams with corresponding location information (lat long coordinates). Datasets from all over the world are welcome. Longer time-series would be great, but also short sampling series could be useful. In case you have large datasets for other pathogens, we would be very happy with those too, as we are expanding our model to include other enteric waterborne pathogens. Do let us know in case you have data that you are willing to share with us (for validation only or for joint publication etc). We will then contact you to discuss the details.
Modelling is a great opportunity to create an understanding of spatial heterogeneity and to identify areas with high concentrations. However, to build trust in our model, observational data are indispensable. We hope you are able to help! Thank you very much in advance for your e-mail!